
The digital universe housing humankind’s combined electronic footprint is expected to jump tenfold between 2013 and 2020, reaching a staggering 44 trillion gigabytes of data—a figure so massive, it's represented by 6.6 stacks of tablets stretched to the moon. Emerging IoT and big data markets are fueling a growth unsustainable in the long term by contemporary storage systems. One major solution developed by researchers from the University of Washington and Microsoft envisions shrinking an entire supermarket worth of data down to the size of a sugar cube by storing it in strings of DNA.
Presenting a paper at April’s ACM International Conference on Architectural Support for Programming Language and Operating Systems, the University’s joint team of computer scientists and electrical engineers detailed one of the first complete systems to encode, store, and retrieve digital information from DNA. The paper also described how the team successfully encoded nucleotide sequences of synthetic DNA snippets with the deconstructed binary data that makes up four digital images. More importantly, the team was able to extract the correct sequence of data needed to reconstruct the images without incurring any data loss.
Compared to existing long-term storage methods like hard drives or magnetic and optical media, DNA molecules can store significantly denser amounts of data, and reliably preserve it for centuries, where the former degrade within a few years. Luis Ceze, one the researchers involved in the project, describes the process as:
“Life has produced this fantastic molecule called DNA that efficiently stores all kinds of information about your genes and how a living system works — it’s very, very compact and very durable. We’re essentially repurposing it to store digital data — pictures, videos, documents — in a manageable way for hundreds or thousands of years.”
To carry out the process, the team needed to develop a method of converting the long binary strings of zeroes and ones into the four basic building blocks of DNA—adenine, guanine, cytosine, and thymine. The ideal logic resided in the Huffman coding algorithm, an entropy encoding method that uses a variable-length code table for lossless data compression, which when applied to DNA molecules, allows digital files to be represented in DNA.
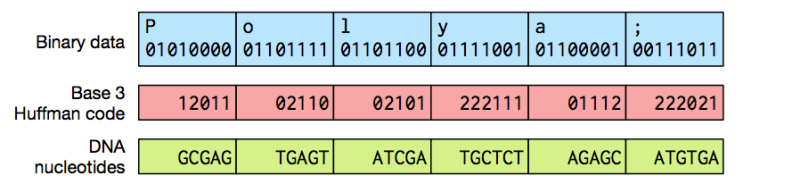
Specific markers are inserted within the DNA strands to allow the files to be retrieved. By applying the Polymerase Chain Reaction (PCR) techniques from molecular biology, scientists can identify a file’s starting point, and recall the specific combination of adenine, guanine, cytosine, and thymine before running Huffman coding algorithm in reverse to reconstruct the file.
Although the technique holds future potential, the current cost attribute of performing the to-and-fro data conversion is unpractical on the larger scale—at least not until the right incentives are in place.
Source: Washington University and Gizmodo
Advertisement
Learn more about Electronic Products Magazine





